By Anderson D. S. Duraes
This is an interdisciplinary project touching on Chemistry, Physics, Mathematics, Computer Science and Engineering.
This work appears in J. Phys. Chem. B 2021, 125, 42, 11709–11716. 10.1021/acs.jpcb.1c07127
and also on ChemRxiv: 10.26434/chemrxiv-2021-196zw
Chemistry
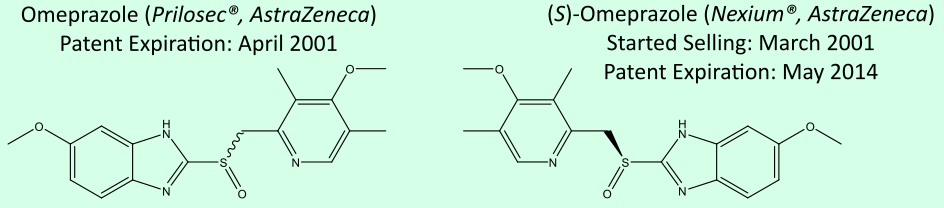
Enantiomers (or chiral molecules) are two compounds with the same structural formula, but which are non-superimposable mirror images. Due to their identical physical and chemical properties in an achiral environments such as water, the separation (or resolution) of the enantiomers is expensive and involves either chiral columns or the use of diasteromers during synthesis.
Physics
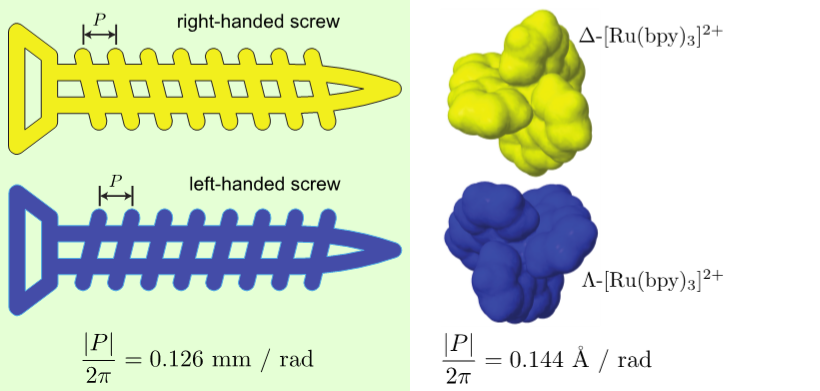
Left- and right-handed screws are also chiral objects that couple rotation around one axis with displacement along that axis. The displacement (in opposite directions) can be measured with a geometric property of the screw: its pitch. In a lab-fixed frame, left- and right-handed screws have pitches with the same magnitude, but with flipped signs, rotating in opposite directions to do the same task.
Under the same counterclockwise rotation, the video shows that the blue (left-handed) screw moves down and the yellow (right-handed) screw moves up.
Mathematics
In this project, we discovered that this geometric property (the pitch of a screw) can be extended to molecules. Enantiomers have molecular pitch values with the same magnitude, but with flipped signs, moving in opposing directions under the same rotation source. As a consequence, this enables the separation of enantiomers.
Computer Science
Significant algorithmic development has gone into both the numerical calculation of molecular pitches and molecular dynamics simulations of solvated enantiomers to verify our predictions. The computed molecular pitches match the molecular dynamics simulations and can be estimated on a time scale of seconds, while the simulations require days or weeks of computer time.
Python code to compute the molecular pitches accompanies the publication and a C++ version of the same code is available as a program in OpenMD. The molecular dynamics simulations were carried out using OpenMD.
| Common Name | Generic Name | CAS Number | Molecular Pitch | P | / 2π (in Å /rad) |
| Aricept | Donepezil | 120014-06-4 | 0.267 |
| Casodex | Bicalutamide | 90357-06-5 | 0.246 |
| Lipitor | Atorvastatin | 134523-00-5 | 0.125 |
| Nexium | Esomeprazole | 119141-88-7 | 0.145 |
| Paxil | Paroxetine | 61869-08-7 | 0.159 |
| Singulair | Montelukast | 158966-92-8 | 0.325 |
| Zoloft | Sertraline | 79617-96-2 | 0.110 |
| (Λ or Δ)-[Ru(bpy)3]2+ | 50525-27-4 | 0.144 | |
| (S)-(−)-1,1′-binaphthyl-2,2′-diamine | 18531-95-8 | 0.222 | |
| CHBrClF | 593-98-6 | 0.0129 |
Engineering
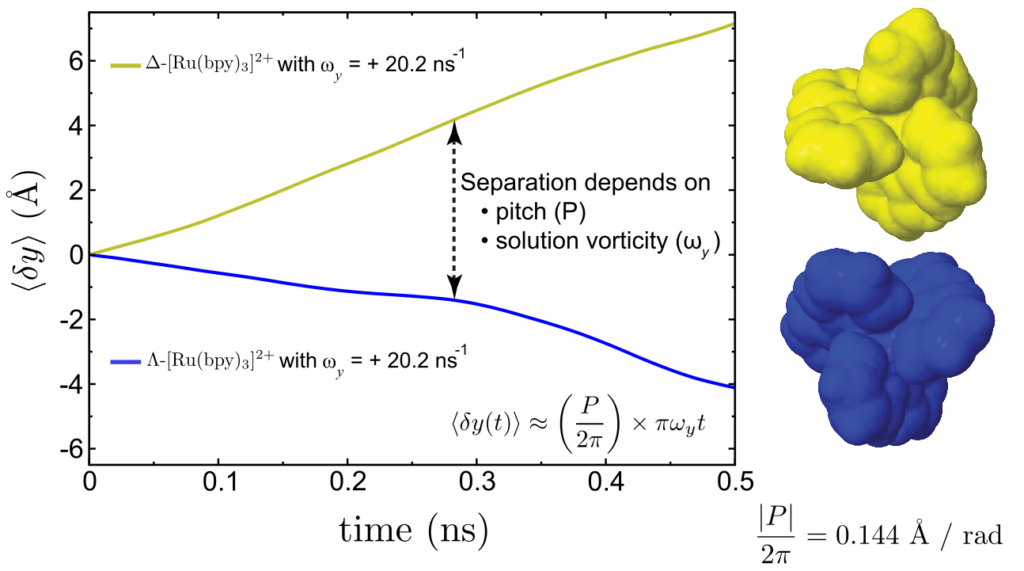
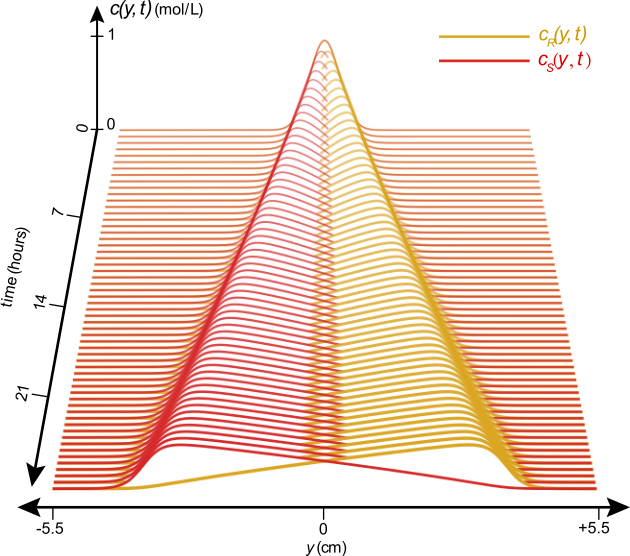
This work also allowed us to suggest design parameters for a device to separate enantiomers on a centimeter length scale using shear flow. This separation can be achieved in hours, when theoretically injecting a 1:1 mixture of enantiomers whose molecular pitch is 0.1 Å / rad in a machine that provides a constant vorticity of 106 s-1. Figure 4 below shows the estimated separation of the concentration profiles as a function of time in a hypothetical Taylor-Couette device.
Other Applications
“Think of the Possibilities!”
(Rupert TV Series, Series ©1995 Nelvana Limited)
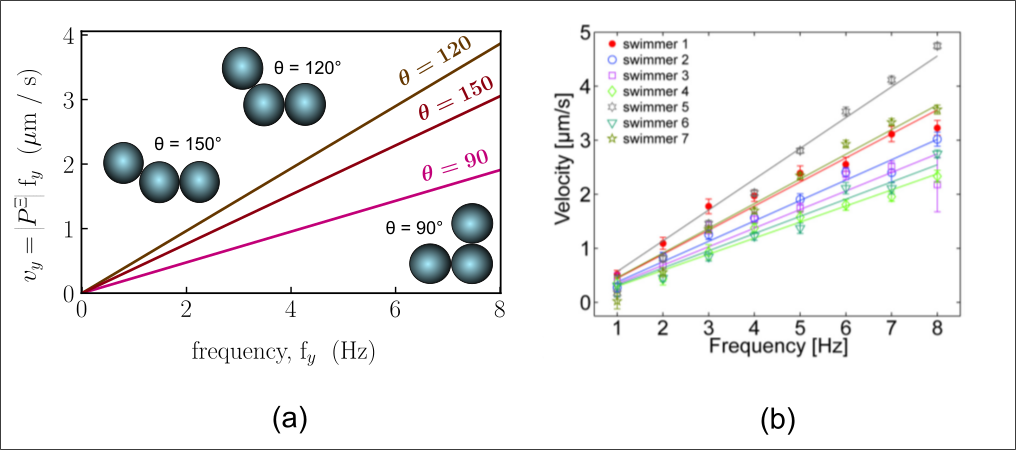
Because our work relies on a first-principle approach, i.e., without data fitting, we can apply the same theory to large objects. For example, we can predict very similar behavior to the experiments on ferromagnetic microparticle ‘swimmers’ that were recently propelled through a stationary fluid using magnetic rotation by Cheang et al.
(Part of the “swimming_far_from_boundary.avi” video in Suplemental Material of Cheang et al.’s paper)
As a final remark, the theory behind this work can be applied to objects of any scale and has the potential to reduce the cost of drug molecules, which are commonly pure enantiomers which currently do not have a general method of separation.
